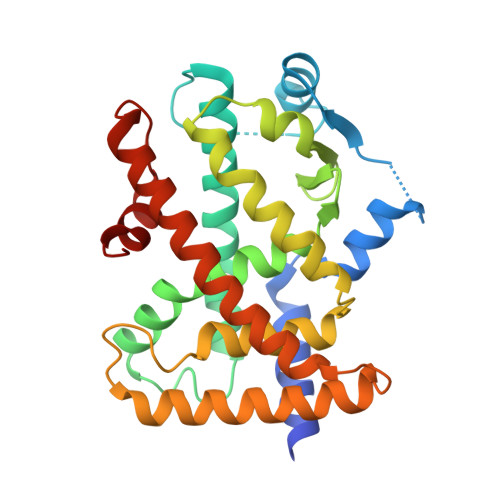
Crystal structures of the ligand-binding domain of human peroxisome proliferator-activated receptor delta in complexes with phenylpropanoic acid derivatives and a pyridine carboxylic acid derivative.
Oyama, T., Takiguchi, K., Miyachi, H.(2022) Acta Crystallogr F Struct Biol Commun 78: 81-87